The Complex Case of Antisense Oligonucleotides

The landscape of oligonucleotides is constantly growing and developing. The increasing complexity of antisense oligonucleotides (ASOs) brings with it a world of possibility in terms of the development of nucleic acid-based medicines and therapeutics, such as RNA-based therapeutics or microRNA therapeutics. Computational developments also open the door to novel tools and technologies for advanced oligo synthesis and impurity analysis.
At Oxford Global’s Oligonucleotide Chemistry and Therapeutics Symposium: Online in 2022, key opinion leaders presented on a range of topics, including oligo chemistry and purification analysis, novel application areas in oligo therapeutics, and innovations in oligo design and delivery for better therapeutic potential. In this insight article, we look at a couple of the best-received presentations given by Lukasz Kielpinski, Principal Scientist at the Roche Innovation Center Copenhagen, and Andre Gerber, Professor at the University of Surrey.
Presentation 1: The Evolution of Computational Representation of Antisense Oligonucleotides
Presented by: LUKASZ KIELPINSKI, Principal Scientist at Therapeutic Modalities, Roche Pharma Research and Early Development, Roche Innovation Center Copenhagen
Nucleic acid-based medicines are continually becoming more complex. Whereas they initially only utilised unmodified DNA, therapies that rely on oligo chemistry and use antisense oligonucleotides with multiple base, sugar, and backbone modifications are being widely developed. With this increasing complexity comes a growing need for accurate methods to represent the compounds used.
Kielpinski began his presentation with an overview of the requirements of an ideal oligo representation system. Firstly, it would include an unambiguous representation of atomic structures, including stereochemistry specification. It would also feature bijection with atomic structure and the ability to represent a broad chemical space, including base, sugar, and backbone modifications, conjugates, and multi-molecular structures such as siRNA. Finally, it should be both human and machine-readable. Kielpinski was careful to note that “these features would be the ideal of oligo representation systems, but they do not necessarily exist at the moment.”
- AI-Discovered Peptide has Dramatic Impact on Muscle Conditioning
- Computational Tools and AI/ML for Antibody Engineering
- MIT Team Create AI-Generated Universal COVID Vaccine
There are several pre-existing methods to represent ASOs. Graphical representations (see Figure 1) come in molecular structures and simplified HELM sketches. Alternatively, symbolic representations, like "Wave" or "Ionis" chiral patterns, and OligoArt, are more straightforward. However, graphical representations can be laborious to write, are inconsistent, and are not machine-readable or searchable.
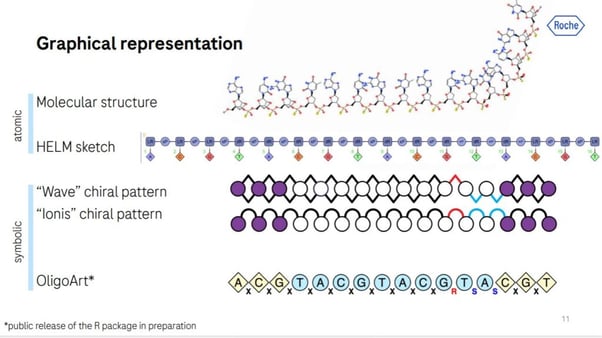
Typographical representation is the most common form of representation and uses a four-letter encoding that reflects uniform modifications via CamelCase or underlining. Multiple modifications are shown with additional styles like italics or colour coding. It's easy to read and write but lacks uniformity between publications and implies a uniform backbone.
Multi-string representation uses run-length encoding or three-string notation, which is easy to manipulate but has a limited character set and does not provide an atomic description. Chain representation, such as concatenated, IDT, and HELM, is machine-readable and offers a full molecular description but is hard for humans to read and requires expertise in defining the monomer library.
HELM, or Hierarchical Editing Language for Macromolecules, is Kielpinski’s preference. It supports describing complex biomolecules of different classes, such as peptides or nucleic acids, and it is expandable to other types of polymers. It also supports chemical modifications, and each monomer has a defined atomic structure and attachment points. HELM is useful for storing information about exact molecular structure because each monomer is defined on an atomic level. However, it is still challenging for humans to read and manipulate.
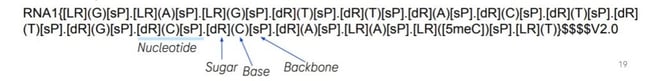
Kielpinski closed his presentation with a discussion about XNAString, an R package that has been publicly released as part of the Bioconductor suite of bioinformatics software. XNAString object defines a compound, base sequence, where the modified bases can also be described, as well as the sugar sequence and the backbone. Once a structure is created, an XNAString object stores information about the compound structure and its conjugates and automatically predicts the secondary structure.
XNAString is closely integrated with HELM notation, meaning it can convert a HELM string into an XNA object. XNAString can also create HELM strings from an XNAString object, making the process far more efficient. Roche is now looking to improve XNAString’s integration with Bioconductor and to improve its usability and performance. Ultimately, XNAString has the potential to make the computational representation of ASOs more uniform and more accessible for both machines and humans.
Presentation 2: The Application of Antisense Oligonucleotides for Purification of RNA-Protein Complexes
Presented by: ANDRE GERBER, Professor of RNA Biology at the University of Surrey
RNA is a vital molecule that performs a variety of functions in cells. RNA-binding proteins (RBPs) are crucial in controlling the fate of RNA molecules in cells, including during transcription, processing, splicing, export, and translation. In addition to RBPs, non-coding RNAs such as miRNAs regulate RNA processing and function. RBPs are also responsible for forming ribonucleoprotein (RNP) complexes. RBPs are defined by their RNA-binding domains, which govern specific interactions with RNA molecules. Based on these domains, around 1400 RBPs have been predicted in humans.
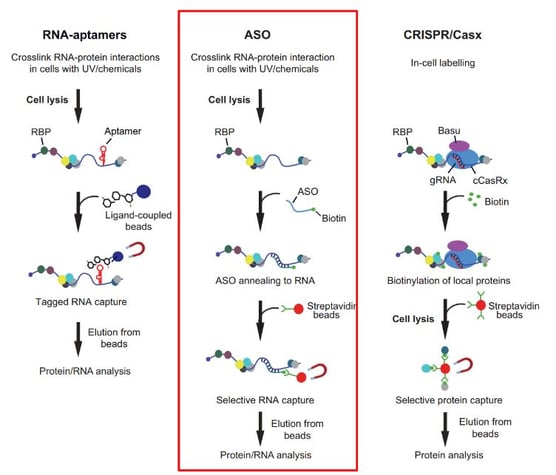
Research in this field is frequently concerned with developing experimental approaches to define RNA protein interactions at a global level; there are several approaches to achieve this, which are commonly referred to as RNA-centric approaches. Gerber’s presentation focused on how ASOs can capture a target RNA sequence. This technique has already been used to target long non-coding RNAs, virus RNAs, and mRNAs.
Fundamentally, an ASO which is complementary to the target sequence is designed and targets a specific RNA molecule within the RNP complex. The ASO is combined with biotin via a flexible linker and conjugated to a solid support, such as magnetic Streptavidin beads, which allows for the capture of the entire RNP complex.
One such approach is TRIP (Tandem RNA Isolation Procedure), which allows for the selective recovery of mRNA and bound proteins (see Figure 3). It uses 21-25-mer 2'O-Met RNA and a 3'-biotin tag with a TEG linker, allowing increased flexibility for increased binding. In Gerber’s procedure, a 2-step protocol called tandem purification is used. First, the poly(A) RNAs are purified with oligo dT magnetic beads, and then modified ASOs are used to capture specific RNAs from the pool via annealing and recovery.
Gerber noted that “RNA target site selection can be challenging due to the need to find an accessible site for the oligo.” To overcome this, structure prediction is broken down into smaller fragments, allowing the selection of accessible regions. Gerber said that researchers typically design three to six oligos across different sites, with balanced AT/GC content and good complexity, which increases specificity. Systematic BLAST searches are performed to check for off-target binding. Adopting a UV cleavable linker region to the ASOs may further allow elution of RNPs by a burst of UV light.
The ASOs used in ASO-based RNA-capture methods need to be empirically tested for efficiency and specificity, another factor which can make this process challenging. Overall, further research in this field will no doubt be beneficial, as it will lead to a clearer understanding of RNA processing and function. In turn, this could lead to the development of novel therapies for diseases caused by RBP mutations, such as neuromuscular diseases, or their aberrant expression or function in cancer or inflammation.
Join Oxford Global’s annual Antibody Engineering 2023: Online event today. This intensive 2-day meeting delves into the latest in antibody engineering and antibody-based therapeutics & a meeting place for experts working within engineering, computational tools and antibody-based therapeutics. Download the agenda to join prominent leaders and scientists as they share new case studies, innovative data, and exciting industry outlooks.