Tackling Tumours In-Vitro with 3D Cell Cultures
Tumour modelling is absolutely vital to oncology. Researchers need to accurately understand how cancers interact with their microenvironment in order to optimise preclinical development of curative drugs. This is why creating more accurate models has such an impact on the progress of cancer therapies.
Chief among the ways in which modelling can be improved is to culture cells in three dimensions rather than two dimensions. 3D cell modelling better recapitulates the in vivo tumour microenvironment. This is due to the similar morphology and slower growth that the 3D environment can provide.
According to Raymond R. Mattingly, Professor &Chair of Pharmacology &Toxicology at The Brody School of Medicine, East Carolina, 3D cancer avatars have the potential to get very close to the in vivo context. When Mattingly was working with Wayne State University of Medicine, his team experimented with the capability of Tissue Architecture and Microenvironment (TAME) models.
Mattingly and his team of researchers used TAME models to investigate the particular indications of premalignant breast cancer transitioning to carcinoma and type 1 neurofibromatosis (NF1).
Mattingly's Lab and Endpoints for Live Cell Assays
To best simulate how tumours grow and spread in the body, it is important that cell models consider various aspects of the tumour microenvironment. The driving cancer cells are not the only tumour cells that need to be modelled. Tumours can present as complex networks of fibroblasts, inflammatory cells, and blood vessels, among others. “For breast cancer in particular, we need to think about myoepithelial cells and fat cells,” said Mattingly.
The assays that Mattingly and his team analysed set out to measure the volume, proliferation, deviation from sphericity, and invasive outgrowths of tumour sites in vitro. This required functional imaging of proteolytic pathways in the tumour microenvironment.
Most of the data that the team collected used confocal microscopy and Z-stack techniques in order to capture images of living cells in real time. Mattingly explained that this can create significant problems with imaging, storage, and analysis, as well as requiring specialist software.
Study one: Breast Cancer
Mattingly first described how they used 3D cell culturing to study breast cancer. In the early stages of cancer metastasis, cancer cells infiltrate into vascular networks, specifically via the lymphatic networks.
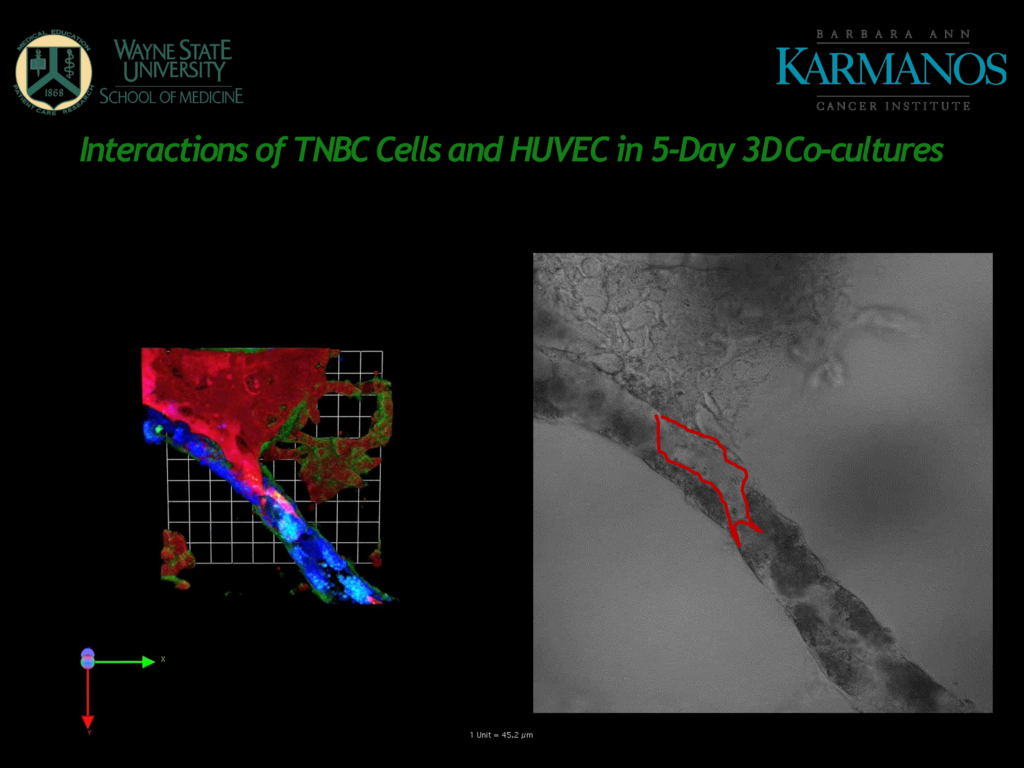
Figure 1 – A slide from Mattingly's presentation
Figure 1 shows human umbilical vein endothelial cells (HUVECs) in blue, which form tubes in the 3D cultured matrix. Cancer cells shown in red can be seen to invade the HUVEC tubes and proteolysis is taking place in the sites shown in green.
“The fact that they model blood vessels rather than lymphatic vessels can be a limitation for the cancer indications that they are studying”
Although HUVECs are widely used for this kind of modelling, Mattingly commented that the fact that they model blood vessels rather than lymphatic vessels can be a limitation for the cancer indications that they are studying. Cancer cells, especially in breast cancer, are more likely to use lymphatics rather than blood vessels as an initial point of entry.
Next, Mattingly showed us a comparison between a model for microvascular blood endothelial cells (BECs) and microvascular lymphatic endothelial cells (LECs).
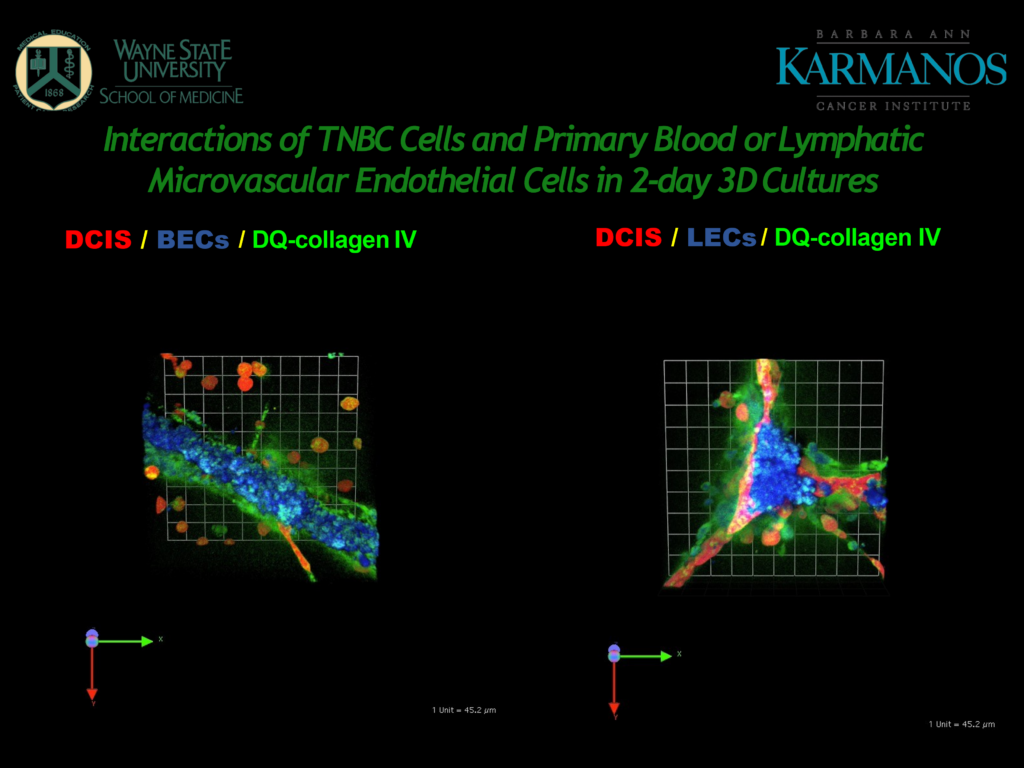
Figure 2 – A slide from Mattingly's presentation, showing 3D cultures of BECs and LECs.
Looking at the BECs (in Figure 2), the cancer cells show some propensity for interaction and there is a lot of proteolysis, “this is associated with the microvascular endothelial cells seemingly even more than with the cancer cells,” said Mattingly.
The LECs on the right-hand side of Figure 2 form in what Mattingly calls “prismatic structures,” which are characteristic of how they would behave in vivo. Mattingly pointed out how the cancer cells have migrated to the LEC structures and there is a large amount of proteolysis.
This led Mattingly's team to coculture LECs and breast cancer cells (specifically MDA-MB-231) together in 3D. The analysis showed that the cancer cells were significantly stimulated by the lymphatic cells and that the lymphatic cells were “massively” stimulated by the cancer cells.
This work, conducted by Mattingly and his team, demonstrates a very significant point. That modelling lymphatic networks in 3D cultures can provide a detailed picture of cancer cells' initial point of infiltration in the body.
This research could be a great leap toward developing new therapeutic endpoints in the future. Drug developers now have a much clearer way of understanding how breast cancer propagates via the body's pathways, certainly an energising prospect.
Study two: Type 1 Neurofibromatosis
Type 1 Neurofibromatosis (NF1) is one of the more common rare disorders - as defined by the EU and US authorities. The condition is punctuated by the growth of large premalignant tumours along the nerves called neurofibromas. Mattingly clarified that “neurofibromas are complex tumours, presenting many different cell types in the tumour microenvironment.”
“Surgery is usually not possible as a curative measure.”
Although neurofibromas are initially benign, they can cause serious problems due to their large size. They can displace organs or other important structures in the body. Furthermore, they also have a high risk of becoming malignant. Due to their complexity, their mass, and their tendency to wrap around vital structures, surgery is usually not possible as a curative measure.
“NF1 is caused by a loss of neurofibromin which is a negative regulator of the Ras proto-oncogene,” Mattingly told us. “When neurofibromin is missing, Ras becomes too active. This induces too much signalling through pathways such as MAPK and PI3 Kinase drives tumour formation, proliferation, and survival.”
This led some researchers to theorise the use of MAPK MEK inhibitors as a therapy for NF1 patients. Although they had promise, Mattingly clarified that disappointingly, there was little selective toxicity in the tumour cells versus normal Schwann cells, which indicated this might not be an effective treatment.
However, Mattingly and his team decided to revisit this idea, this time using 3D cell cultures to model the inhibitors' effect. They analysed the influence of the cell inhibitor selumetinib on the neurofibroma microenvironment and compared their analysis using both 2D and 3D cultures. This comparison hoped to reveal whether the 3D environment would better elucidate the drug's effect. “Truly pathomimetic 3D cultures should be suitable enough to identify the driving pathways that are sensitive to targeted inhibition," stated Mattingly.
They found that there was a suitable amount of selectivity in their 3D models to serve as evidence for some neurofibromas' sensitivity to the MEK inhibitor. Other teams studying the effect of MEK inhibition in inoperable neurofibromas concurred with this conclusion. Their results found that around half of the paediatric patients saw significant tumour shrinkage with selumetinib.
The drug has now received approval for the treatment of NF1 and is the first drug on the market of its kind. Unfortunately, selumetinib does not treat all cases, so the team are still studying plexiform neurofibromas. Mattingly explained that his team are now “interested in the potential of high-content analysis to think about proteolysis as a problem.”
The advent of 3D cell modelling brings abundant opportunities for drug discovery and development, as demonstrated by Mattingly's research. Bringing an extra dimension to models could indeed expose new horizons of dimensions to therapeutic modalities. Having climbed one mountain, the drug discoverer is now akin to the wanderer above the sea of fog.
Our 3D Cell Culture congress incorporates key trends and innovative technologies to accelerate the adoption of 3D models in preclinical research via advanced development, validation, and application strategies.