Spatial Transcriptomics: Innovations & Novel Tools
With the development of single-cell RNA sequencing, it became possible to measure transcriptomes at a single-cell resolution. This was a considerable achievement for the scientific community. However, despite its impact, single-cell RNA sequencing has some limitations. Tissues are typically dissociated into single cells, and the cells are taken away from their context within the tissue. Unfortunately, the majority of the transcriptome-wide spatial assays do not operate on a single-cell level. Instead, they produce data comprised of contributions from a – potentially heterogeneous – mixture of cells.
Recent advances have pushed single-cell transcriptomics further than basic profiling with RNA-seq. Innovative approaches can capture multiple characteristics of a single cell’s immunophenotype, genome sequence, lineage origins, DNA methylation landscape, chromatin accessibility, and spatial positioning. However, each technology has unique strengths and weaknesses. In addition, to date, there is no all-encompassing method, creating the need to combine and integrate datasets from disparate tools.
Innovative Methods and Tools
Several tools and methods have been created to improve or combine single-cell RNA sequencing and spatial transcriptomics data. Progress has been rapid in the last few years; hundreds have tools have been designed to tackle issues and challenges as they have developed. While this is undoubtedly a good thing, it has led to fragmentation and difficulties integrating data parsed with different software.
Giotto Package
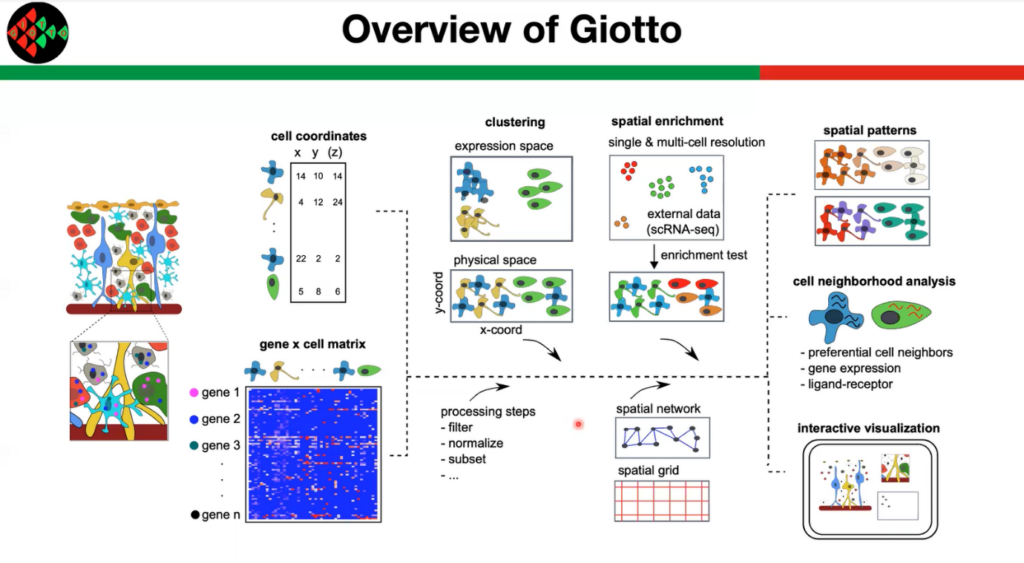
Giotto is a tool designed to analyse and visualise spatial expression data. Guo-Cheng Yuan (Professor at Icahn School of Medicine) explains that technologies “such as the Visium and Nanostring are great, but one of the limitations is that they do not yet have a single-cell resolution. You want to know the cell type distribution. So, we developed Giotti to do cell type enrichment analysis. The basic idea is to take the cell-type-specific gene expression information from the external single-cell RNA-seq data and look for the enrichment across tissues. And the enrichment is going to tell you the distribution of the different cell types.”
Giotto recently demonstrated applicability on nine different techniques, including in situ hybridisation, sequencing, image-based multiplexing, and proteomics. Wide-ranging applicability is at the core of Giotto’s design, allowing adjustments for single and multicell resolution and both 2d and 3d spatial dimensions, varied throughput and different numbers of cells and genes. Guo-Cheng argues that “by combining spatial domain and cell type data, we can start to understand the differences between intrinsic and environmentally induced variability. Data input has two major components, one is the locations of individual cells, and the other is the gene expression matrix. And we believe that by using these, we can analyse many different data types because these are the common ingredients of all data types. Starting from that, Giotto can do many functions starting with clustering, we can also do spatial enrichment, and we can look at other spatial networks. From that, we can look at the spatial patterns, the cell neighbourhood analysis, and the cell-cell interactions. And finally, we can have a visualisation tool that enables users to explore the data interactively.”
Final Thoughts & Conclusion
The advancement and integration of single-cell and spatial transcriptomic methods are one of the most exciting areas for academic institutions and pharmaceutical companies. This year we will hold three in-person spatial conferences in the UK (27 - 28 April 2022), Germany (07 - 08 June 2022) and the USA (September 2022). If you are interested in spatial technology and methods, we hope you will be able to join us in person or at one of our upcoming virtual discussion groups.