Single Cell Proteomics: Sample Preparation and Tissue Selection
Proteomics has faced many challenges for application at single-cell resolution. Other omics, such as genomics and transcriptomics, can use amplification to aid single-cell analysis. Unfortunately, proteins cannot be amplified, meaning that research is limited by the small number usable in a single cell. Proteins provide unique insight into cell structure and processes. Furthering single-cell proteomics is a top priority for many researchers and pharmaceutical companies.
Single-cell proteomics has developed quickly, despite being a relatively new field. Until the 2010s, protein levels in single cells were estimated using cellular mRNA levels or multi-cell samples. Fortunately, novel proteomic approaches hold the promise of direct testing and new insights. Single-cell proteomics aims to observe protein interaction and changes directly, enabling a more complete understanding of cellular activities.
Miniaturisation and Sample Preparation: NanoPOTS
At Oxford Global’s 2022 single-cell proteomics workshop, Ryan Kelly, Associate Professor in the Department of Chemistry and Biochemistry at Brigham Young University, recounted his team’s work on NanoPOTS. His work aimed to minimise exposed surfaces and, therefore, nonspecific adsorptive losses through sample miniaturisation. He explains that “the approach that we hit on was basically keeping the benefits and the general form factor of conventional workflows, and just miniaturising it to reduce surface exposure volumes and, also incorporating a one pot sample preparation workflow. The acronym NanoPOTS stands for nano-droplet processing in one-pot for trace samples.”
The team recruited Ying Zhu to help with this. He had previously been developing a robotic nano pipetting platform. While his previous work involved using an oil cover, the new approach involved using a humidifying chamber for droplet dispensing. Kelly clarifies that this approach means that they are “only ever adding a reagent, incubating, and then adding the next reagent, so it all takes place in the same well.” While the initial publication did not get down to the single-cell level, the team achieved a proteome coverage of over 3000 protein groups in approximately ten cells, around 500 times more than any previous publication.
Single Cell Analysis: NanoPOTS and FACS
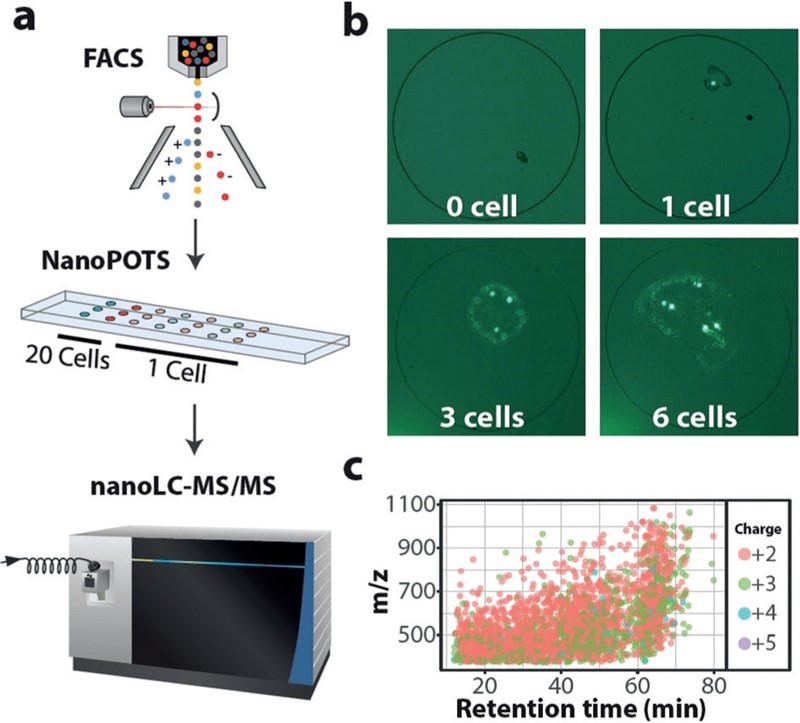
Shortly after their initial publication, Kelly’s group combined the NanoPOTS platform with fluorescence-activated cell sorting (FACS). They aimed to evaluate the potential of NanoPOTS for analysing proteins within single mammalian cells. He claims that “this works well if you have the right FACS system. I think for making spatial measurements, coupling with laser microdissection is also very valuable.”
Using FACS in tandem with NanoPOTS offers three main advantages. First, it allows far more precision when loading cells into each nanowell. Second, FACS reduces background contamination via cell suspension in a phosphate-buffered saline (PBS) solution. Third, FACS provides vital visualisation for biochemical analyses, allowing greater detection and isolation of specific cell types.
Based on MS/MS identifications, researchers initially identified 200 to 300 protein groups from 50 micrometres of material. Since then, advancements have pushed this number up significantly while leaving additional room for improvement.
Tissue Selection: Fresh Frozen or Formalin-Fixed Paraffin-Embedded?
Another essential contributor to the success of single-cell proteomic analysis is tissue selection. Fresh frozen has is favoured in omics sciences due to higher quality preservation, but it’s generally more expensive. Kelly explains that “most of our tissue uses frozen tissue. There are vast resources of archived FFPE tissues and they may hold the answers to many biological questions. And so, more recently, we have been working with FFPE tissues and looking at applying both nanowell-based prep and fully automated, well plate-based workflow that we call autoPOT.”
While the quality of results gathered from FFPE are not as impressive as with fresh frozen, adjustments can be made to improve them. Kelly shares his experience, noting that “if we use the same conditions that we use for fresh frozen tissue, we take a pretty big hit on coverage. But it turned out if we just increase the temperature and duration of incubation, we get to about 85% of the coverage for an equivalent tissue. And this truly is equivalent tissue, we just recently tested this using a mouse liver with half preserved by FFPE and half by fresh freezing.”
Conclusion and Final Thoughts
Sample preparation and tissue selection are critical to successful single-cell proteomic analysis. However, challenges remain before single-cell methods can become as common as other forms of analysis. Fortunately, leading researchers such as Kelly and his team are hard at work designing new single-cell proteomics methods.
For the most up to date information on single-cell proteomics, consider becoming an Omics Community member. Becoming a member provides access to our events as well as on-demand presentations, including Kelly’s full one hour workshop.