Quantitative Mass Spectrometry: a Clinical Tool for Biomarker Discovery and Qualification
Presented by: Axel Ducret, Senior Principal Scientist at Roche
Edited by: Ben Norris
Many promising protein biomarkers discovered and qualified in pre-clinical models are typically not followed up in the clinic. At the same time, proteomics – done through quantitative mass spectrometry – represents a time and cost-effective means of building assays suitable for testing hypotheses in multiple settings, including in clinical trials. For Axel Ducret, Senior Principal Scientist at Roche, clinical proteomics means the opportunity to analyse relevant human clinical samples, as well as contributing to critical aspects of biomarker strategies.
An Overview of Quantitative Mass Spectrometry
There are several features of mass spectrometry that make it appealing for biomarker discovery and qualification, as Ducret explained to our Biomarkers Week: Online in 2021. As an antibody-free method, it saves time and money through the direct, specific detection of an analyte. The instruments involved are highly quantitative, especially with the use of internal standards, and the dynamic range is very large. “What is most exciting about mass spectrometry is the multiplexing capability inherent to the method,” said Ducret. “Typically, tens of proteins can be monitored in the simultaneously targeted approach, and up to several thousands can be monitored in an unbiased approach.”
“Nowadays, even the minute amounts of samples collected in clinical trials can be analysed by the highly sensitive instruments we have at our disposal.”
Significant advances have been made in mass spectrometry regarding the analysis of biofluids and formalin-fixed paraffin-embedded (FFPE) tissue. “Nowadays, even the minute amounts of samples collected in clinical trials can be analysed by the highly sensitive mass spectrometry instruments that we have at our disposal,” said Ducret. He added that quantitative mass spectrometry workflows in biofluids and FFPE samples are being developed and validated, with several guidelines now in place to regulate sampling procedures and assay qualification.
Protein Biomarkers in FFPE Tissue
Having established some background for the functionality of mass spectrometry-based assays, Ducret moved on to discuss a proof-of-concept study from 2014. He explained that the study was carried out to demonstrate that FFPE tissue was suitable for the measurement of biomarkers. A targeted assay was developed on a well-established biomarker – in this instance, HER2 – to be measured in breast tumours. Results of the mass spectrometry-based measurements were to be matched with data obtained using standardised assays such as immunohistochemistry (IHC) or fluorescence in situ hybridisation (FISH). These two diagnostic methods are routinely used to determine HER2 levels in tissue and trigger patient eligibility to an anti-HER2 treatment in the case of overexpression.
- Making Mass Data Make Sense: Spatial Multi-Omics and Cytometry
- Discussion Group Report: Spatial Biomarkers and Multiplexed Immunofluorescence
- Human Stem Cell Models of Neurodegeneration
The generic proteomics workflow for FFPE tissue samples starts with a formalin-fixed tissue block embedded in paraffin. The block is sectioned in slices of five to ten micrometres thick, with one of them stained with haematoxylin and eosin (H&E) to localise the tumour. The remaining unstained sections are de-paraffinated and rehydrated, and the tumour is macro-dissected and put into a tube. After antigen retrieval, proteins are trypsinised and the generated peptides are collected and subsequently analysed via a mass spectrometry workflow.
In this study, results obtained from targeted proteomics agreed well with IHC findings and only in part with FISH, reflecting most likely differences in protein expression versus gene amplification. “Having convinced ourselves that mass spectrometry can be used for protein quantification in this setting, we embarked on a follow-up project,” said Ducret. “In this second project, our aim was to investigate whether it was possible to build a highly multiplied assay on the basis of a list of proteins of interest.”
Assay Development for Triple Negative Breast Cancer Identification
For this second project, Ducret and his team investigated triple-negative breast cancer (TNBC), which represents an important clinical challenge. It accounts for between 10-15% of all breast cancers, and is termed triple-negative because cells of this cancer type don’t display oestrogen or progesterone receptors and return negatives for HER2 overexpression. The high risk of relapse and poorer prognosis, as well as the quick rate of growth, makes TNBCs a continued difficulty to identify and treat.
Ducret explained that his team aimed to develop a multiplexed, targeted mass spectrometry assay panel starting from TNBC-related targets identified from various sources, including genomics and proteomics studies, to enable a future quantification of proteins in a cohort of TNBC tumours. In this study, a list comprising 374 protein candidates was first qualified for their suitability to be included in a targeted mass spectrometry assay, after which 224 candidates underwent assay development resulting in a panel of 200 proteins being assayed in two liquid chromatography-mass spectrometry (LC-MS) analyses.
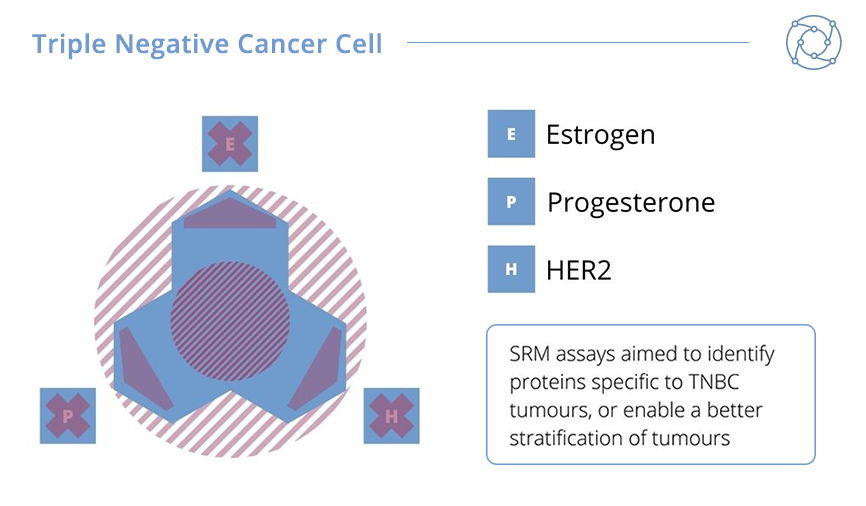
The results from Ducret’s TNBC biomarker project were promising. A sample tumour panel comprised of 20 HER2-overexpressing tumours – including Luminal B tumours – and 20 Luminal A tumours were compared against 49 TNBC tumours. Ducret explained that 185 of 200 qualified proteins could be measured in the panel. HER2-overexpressing and Luminal B tumours were shown to have an increased expression of HER2 protein whilst elevated levels of the oestrogen and progesterone receptors were measured in Luminal A and B tumours. As expected, TNBC tumours were characterised by an absence or very low levels of any of these markers. In addition, Ducret announced that seven novel biomarkers apparently specific for TNBC tumours were selected for additional investigation in future studies.
Areas of Focus for Future Clinical Proteomics Analysis
“For me, it is quite obvious that clinical proteomics is becoming a reality,” said Ducret to summarise the presentation. Comprehensive proteomics coverage with accurate quantification will be key to developing effective techniques for the identification and targeting of threats such as TNBC tumours in years to come. “Now, of course, a clinical proteomic study needs to be prepared and carefully executed,” added Ducret. “As for any study, the goal needs to be clearly defined ahead of time.”
Well-characterised clinical samples should be in sufficient abundance for future research in order to maintain reliable selection and surveying criteria. Biomarker discovery in biofluids is dependent on a strict adherence to the sample collection protocol. “I am confident that in a few years such approaches will be much more commonly-used, and will provide critical information to better treat our patients,” Ducret concluded.
Want to learn more about the latest innovations in the Biomarkers field? Visit our dedicated portal for cutting-edge industry insights and in-depth features. If you’d like to register your interest in our upcoming Biomarkers UK: In-Person event, click here.