PROTACs: “Exciting Opportunities to Explore New Biology”
Rick Davies is the Senior Director of Chemical Biology and Protein Science at Discovery Sciences, a cohort within AstraZeneca that focuses on early drug discovery. “Omics and other data that is now becoming available is beginning to potentially revolutionise our approach to identifying new targets and increasing the number of new targets that are identified,” said Davies at Oxford Global’s 2021 Discovery week.
He pointed out the outstanding wealth of next generation therapeutics that have joined the market in recent years, but he specifically zeroed in on the landscape of PROTAC research as his area of interest. Davies’s team have set out create a platform which synthesises PROTACs against targets where potent ligands are available.
One target that Davies’s team considered was IRAK3, an enzyme that suppresses proinflammatory signalling in leukocytes. They were able to take advantage of a non-functional IRAK3 binding ligand by attaching it to a cereblon (CRBN) warhead, to form a PROTAC that targeted IRAK3.
Davies said that the resulting PROTAC was able to “very rapidly induce selective degradation of the IRAK3 protein.” Using global proteomics, they also found that the degradation of IRAK3 was “fairly specific, occurring in a relevant cell line,” which proved the efficacy of their methodology.
Automated PROTAC Synthesis
Now that they had managed the proof-of-concept, it was time to scale up their research. What other targets might the team at AstraZeneca be able to extend this technology to? Davies’s team developed an automated PROTAC synthesis platform.
They created a toolbox, complete with a varied bank of protein of interest ligands and E3 warheads. The warheads they considered were specifically VHL and CRBN, but it is worth noting that the toolbox is future compatible, with a view toward expanding its range of E3 ligases.
Their technology’s modular construction allows for synthesis and purification of the PROTACs that they create to be automated: “It allows us to produce tens to hundreds of PROTACs literally in days.” The team can then swap out ligands, linkers, and warheads, and “very rapidly” test whether the PROTACs that they create are worth continuing to future experiments.
“The next part of the jigsaw is to set up degrader assays that quantify the degradation of these targets,” continued Davies, explaining that there are two high throughput degradation assays that they are working with: an antibody-based and a luciferase-based approach.
Navigating a Plethora of E3 Ligases
Which E3 ligases have the potential to act as therapeutically useful PROTACs? Davies and the team wanted to focus on E3 ligases that had four specific properties: ubiquity, disease specificity, cell-type specificity, and tissue specificity.
With the criteria for what would make a successful PROTAC ligase defined, this led them to develop a way of assessing new E3 ligases. Their system used a dummy target protein joined to a NanoLuc for identification to which they introduced E3 ligases coupled to a halo tag (see Fig. 1).
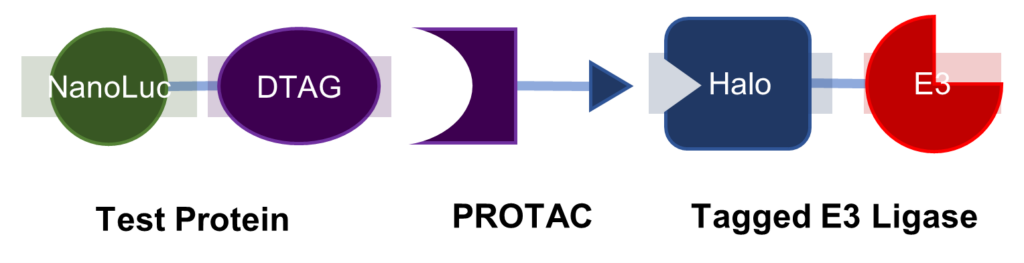
Figure 1 – A diagram showing the system for assessing E3 ligases.
Then the team could construct a PROTAC that consisted of a DTAG ligand and halo ligand, bringing together the test protein and the tagged E3 ligase. Here, Davies and his team could compare those ligases to the traditional ligases such as CRBN or VHL. The results from these experiments validated the E3 ligases for further testing. It showed that the ligases studied were indeed capable of degrading and ubiquitinating targets.
Biological PROTACs
Discovery Sciences has additionally been investigating the potential of biological PROTACs. Davies explains that these are PROTACs in which the protein of interest is targeted by a protein that is encoded onto DNA or mRNA along with the degrading module. As a result, the nucleic acid can be transfected into different cells to produce a bifunctional molecule capable of targeting a specific protein for proteasomal degradation.
Like the previously described synthesis system, Davies’s lab has developed a modular method of synthesising these biological PROTACs. They have created a toolbox of various protein of interest binding domains, linkers, and degradation domains, allowing the molecules to be created using these modular building blocks.
The example that Davies gave was a biological PROTAC that they created to target SHP2 as its protein of interest. The PROTAC involves a VHH coupled to VHL E3 ligase via a linker (see fig. 2) and provided results that strongly suggested the degradation of SHP2 when observed using imaging assays.
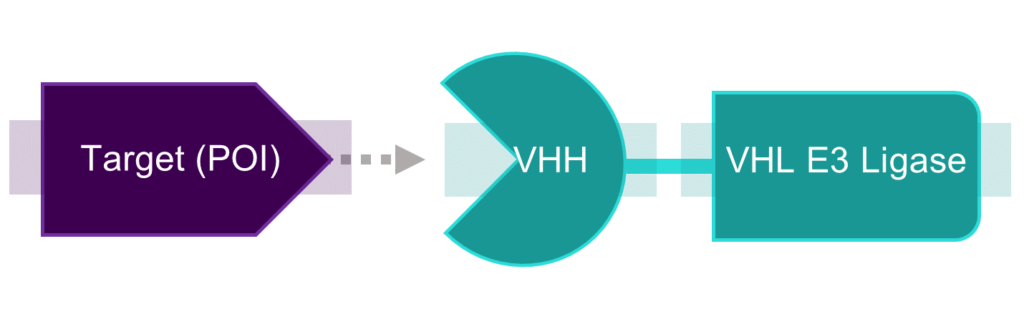
Figure 2 – Diagram representing Davies’s team’s bioPROTAC in cyan.
The next item in the toolbox for the AstraZeneca team to develop was the small-molecule warheads used against the proteins of interest. These were identified using DNA-encoded library (DEL) screening. Then they generated the off-DNA compounds against specific E3 ligases before being attached to the other end of the toolbox using a known POI ligand.
This ‘assessment and tool provision’ allows the researchers to determine the effectiveness of the degradation using a variety of E3 ligases. Davies further noted the potential to run other affinity screenings in future tests, such as HTS and fragments.
Davies and the team are now working to use the methods described to further build their toolbox to effectively degrade a number of different targets, concluding that “PROTACs present exciting opportunities to explore new biology, but they also present new challenges for biology and chemistry alike.”
Join and network with over 400 industry leaders at the renowned Drug Discovery Summit in Berlin, where we will address the latest advancements in target identification, validation and HIT optimisation.




.png)

