Multi-Tissue Molecular Profiling: Precision Medicine for Neurodegenerative Diseases

By Carlos Cruchaga, Professor of Psychiatry, Neurology, and Genetics at Washington University School of Medicine
Transcribed by Tia Byer
Genome-wide association studies (GWAS) for Alzheimer’s disease have so far successfully identified genetic regions associated with neurodegenerative complex traits. However, GWAS have two main challenges. The first problem is that the effect size of the signal is small, reflecting and explaining less than 10% of the complex trait. Secondly, for most signals, it remains difficult to determine whether the signal represents the functional gene the mechanism by which those genes lead to disease.
The Curchaga lab at the Washington University School of Medicine has been performing deep molecular profiling of human samples to understand the biology of Alzheimer’s disease and identify novel causal cell proteins and druggable targets. The platform aims to determine biomarkers and generate novel prediction models for Alzheimer’s to determine the biological context of signals. In particular, multi-tissue and multi-omic data distinguish novel protective variants and drug repositioning in the disease-relevant tissues such as the brain, plasma, cerebrospinal fluid (CSF), and induced pluripotent stem cells (iPSC).
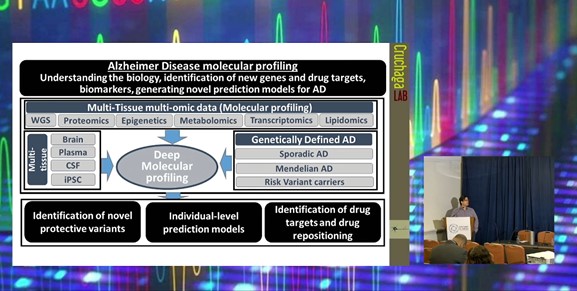
Multi-Tissue High-Throughput Proteomics:
In a study looking at the proteomics in multiple tissues, the Cruchaga lab conducted high-throughput analysis of approximately 1,300 CSF, 650 plasmas, and 450 brain samples. The latter also included lipidomic, metabolomics, transcriptomics, and methylation study. With the data, the lab conducted protein quantitative trait loci (pQTL) and colocalisation followed by Mendelian Randomisation to facilitate identification of causal proteins and druggable targets.
- NGS Data Analysis and Utility
- Integrated Cellular Atlas of Primary Human Breast Cancer Webinar
- Simultaneous, High-Yield Extraction of DNA & RNA from FFPE Tissue
Overall, the lab was able to identify hundreds of novel and unknown pQTLs, 95% of which were then replicated in independent data sets. Whilst most of the novel PQTLs were found in CSF (274) and in plasma (127), only 32 were identified in the brain sample. Results attested to the value of signal identification for GWAS.
Genetic Architecture Comparison:
Comparative genetic architecture analysis is necessary to determine why each tissue subset differs in their tissue-specific signals. To test this, the Cruchaga lab separated the study into cis-pQTL and trans-pQTL. Cis-pQTL signals are those that are close to the gene, qualifying the protein and trans-pQTLs are those that are far away from the gene. Results showed significant tissue-specific signals, with approximately 25% of cis-pQTLs demonstrating tissue-specific signalling and 75% shared signals.
In comparison, trans-pQTLs were shown to be approximately 75% tissue-specific in their signalling. These statistical conclusions provide critical insight into identifying brain and CSF samples as optimal neurological traits for tissue selection. Findings there then incorporated into a multi-tissue pQTL mapping process, to action mendelian randomisation and colocalisation for drug repositioning.
The lab was able to identify hundreds of novel and unknown pQTLs, 95% of which were then replicated in independent data sets.
Creation of Prediction Modes:
The second stage of the study involved testing differential protein levels to discover new prediction models and pathways implicated in Alzheimer’s disease. Researchers then used additional independent datasets to replicate their findings. The Cruchaga lab was able to generate and validate proteomic-based prediction model in CSF and plasma that performed better than the gold standard: CSF Aβ/tau ratio.
Conclusion and Findings:
Overall, the Cruchaga lab at the Washington University School of Medicine identified hundreds of novel pQTLs, the majority of which were in CSF and were replicated in independent datasets. Cis-pQTL but not trans-pQTLs were found to be shared across tissues. However, a large proportion of pQTLs (40 to 70%) are not QTL for other omic modalities. Furthermore, the lab discovered that mendelian randomisation of the pQTL identifies novel proteins involved in complex traits and potential drug targets.
Molecular profiling provides significant understanding of complex diseases.
The lab concluded that molecular profiling provides significant understanding of complex diseases. Transactional data such as genetics and transcriptomics enables discovery of the biological context of neurodegenerative diseases and thorough comprehension of pathological events. Looking ahead, deep molecular phenotyping of clinically characterised cohorts will lead to identification of novel genes and pathways implicated by diseases, novel prediction models, and novel therapeutic targets. Finally, multi-omic data will facilitate a more personalised disease risk prediction and treatment.
Want to hear more about multi-tissue molecular profiling? Join leaders, experts and researchers at our Spatial Biology Europe: In-Person event. The Oxford Global conference will connect global academic & research organisations as well as pharma representatives for high-level discussions on the latest innovations in spatial research & technologies.