GOAL: A Collaborative Approach to Next Generation Sequencing in Oncology and Beyond

Presented by Jeremy Segal, Director of Molecular and Cytogenetic Pathology at the University of Chicago
Edited by Ben Norris
Some of the biggest scientific breakthroughs have been collaborative efforts that stand testament to the strengths of cooperative working. One such success story is the Genomics Organisation for Academic Laboratories (GOAL). GOAL was set up to circumvent some of the traditional barriers to the development of next generation sequencing (NGS) oncology diagnostics at academic medical centres in the US. As Jeremy Segal, Director of Molecular and Cytogenetic Pathology at the University of Chicago explained, collaborative working between academic centres can help to force new ground in improving access to NGS techniques.
Hurdles to Applying NGS in the Clinical Space
“Trying to develop NGS in the clinical space is a challenging process,” Segal told the audience at our 2022 Biomarkers UK: In-Person event. “There’s a lot of different equipment involved – bioinformatics is a big hurdle for all of us, and producing these tests takes a long time in terms of development.” Regulatory challenges can provide a further headache in getting new tests off the ground: prior to the advent of GOAL most academic labs have traditionally worked independently rather than collaborating on NGS methods.
In 2016, Segal and his team were working on a large-scale panel looking at roughly 1,200 genes from solid tumours. “We were putting it together so that it could look for mutations and a variety of genes,” he said. “To make this work, typically if you’re doing very large-scale NGS, you’re usually talking about hybrid capture sequencing.” Segal explained that this involves fragmenting a DNA sample down to make a genomic library. “If there’s a particular fragment that you want to sequence, then you have to make a hybrid capture probe pool and pull down the sequences that you want.”
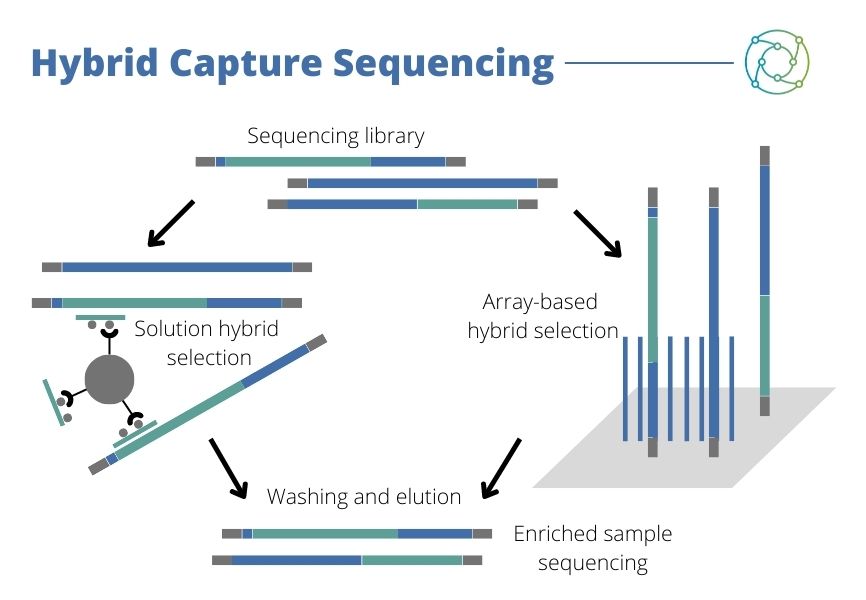
While this might be relatively straightforward on paper, in practice implementing this approach can be much more difficult. “We would get mostly good results, but you’d find some areas that were really problematic,” Segal explained. One example he gave was CEBPA, an important gene for acute myeloid leukaemia. The initial approach involved using large numbers of probes made as part of a batch process, which ran into problems on account of the unreliability of some probes when produced at scale. It became clear a different approach was needed.
Oligonucleotide Probes: Augmenting Analytical Coverage
The main problem Segal’s group was having was generating adequate sequencing data for regions with high guanine-cytosine (GC) content. Efforts to improve performance of their original bulk-generated capture technology repeatedly failed.
“One day we started talking to a vendor from IDT, which is a company which makes oligonucleotides,” said Segal. “They said ‘no, this is a piece of cake’. We had been conditioned to think that capturing and sequencing high GC DNA was difficult to do so we were sceptical, but we were also desperate so we tried it.” The results were tremendous: with the new probes for spiking genes into their reaction, Segal and his team at the University of Chicago were able to achieve ideal sequencing coverage across all previously problematic regions, and their panel development was rescued.
- Tumour Profiling Solutions to Enable Precision Oncology
- What's the Current State of Omics-Based Biomarkers?
- Quantitative Mass Spectrometry: a Clinical Tool for Biomarker Discovery and Qualification
It turned out that the root of the problem was not inherent difficulty in capture or sequencing of high GC DNA, but in the proper manufacturing of high GC capture probes. By synthesising probes individually and performing quality control for each probe, IDT was able to generate far better results.
With the success of this initial pilot, the question of purchasing the entire panel through this vendor was then raised. The manufacturer proposed a very reasonable deal, although this still came to roughly USD $300,000-$500,000 in purchase. “Usually, we’d be paying USD $40,000 per 1,000 samples,” continued Segal. “But then we asked what we’d actually get if we paid this larger amount.” It transpired that the investment would be enough to test 30 million samples, which was a significant return. “For us that’s ten thousand years of tumour sequencing,” said Segal. “So we started thinking, maybe we should try to share this with somebody and buy this as a group.”
Shot on GOAL: Collaborative Purchases for Improved NGS Access
Segal reached out to long-time colleague Dara Aisner from the University of Colorado regarding a collaborative purchase. They had hoped to get three other organisations interested, but they were unprepared for the response they received. “We were really overwhelmed when 15 labs were interested in doing this,” said Segal. While the collective interest of different organisations made the purchase incredibly feasible, it also posed new complications. “With 17 academic centres, there were big content decisions to think about,” Segal continued. “There were a total of 2,640 genes that people were interested in, but over half of them were genes that only one centre cared about. So what do we do?”
In the end, the group decided to buy everything. “We came up with the idea of doing this in a modular format, which allows each site to formulate their own panel or a variety of different panels,” said Segal. “IDT was a really nice partner for this, because you’re talking about buying 95,109 probes divided into 2,670 pools.” There were concerns that the purchase would go awry, but the panel was a success, explained Segal. “We reformulated our panel to about 1,000 genes, and compared with our original panel we had a really nice comparison of all of our variant findings in varying frequencies. So we didn’t embarrass ourselves by wasting everyone’s money and we could show our faces in public. It was great.”
GOAL, Academic Partnerships, and Future Aspirations
Now, 29 academic centres across the United States have access to these probes and work with them every day for clinical services or the development of their operations. “Our expectation was that people would go back to working independently after everyone got the probes, but that’s not what happened,” said Segal. Researchers started asking for advice on using the probes and sequencing methods, with the overall result that research between academic centres became much more cooperative.
“If we’re successful overall, we’ll have an ability to help expand patient access to personalised genomic testing.”
Eventually, it was decided to turn this partnership into a functional organisation – GOAL – with a mission of driving the advancement of cutting-edge genomic testing, specifically by academic and non-academic laboratories. In 2020, Segal and Aisner partnered with the Association of Pathology Chairs (APC) to create a formal non-profit organisation with bylaws and a Board of Managers, and began the process of formalising legal master agreements with the participating institutions.
GOAL is now working on a number of initiatives designed to accelerate testing, reduce barriers and maximize quality across the network. For example, a network-wide concordance is under way to assess lab performance and investigate the potential benefits of implementing a cloud-based consensus bioinformatics pipeline. The group is also laying the groundwork for large-scale sharing of genetic variants and interpretations to assist with review and curation of rare and novel genetic variants in order to enable more rapid scale-up of future sequencing panels.
“If we’re successful overall, we’ll have an ability to help expand patient access to personalised genomic testing,” added Segal. “We’ll get more biomarkers installed in more places, and we can actually help with future biomarker discovery as well.”
Want to learn more about the latest advances in biomarker discovery and sequencing? Head over to our Biomarkers portal for insights from the industry's best and brightest. If you'd like to register your interest in our upcoming Biomarkers US: In-Person conference, visit our event website.