AlphaFold and Drug Design: Has AI Solved Biology’s ‘Grand Challenge?’

The world of medicine has been stumped by a ‘grand challenge’ as influential as it is notorious. Protein folding has for decades been considered enigmatic. The precise way a protein is folded determines much of its functionality within the body, which means mapping a protein’s shape is vital to understanding the drugs that target it.
Scientists have long been able to determine the chemical composition of many proteins. Still, their shape is much trickier since there are an astronomical number of structural configurations in which any particular protein can fold.
- Merck KGaA: Accelerating Drug Discovery with Automation and Artificial Intelligence
- Pharma Manufacturing: Digitalization and Industry 4.0
Enter AlphaFold
The protein folding problem had become such a holy grail for researchers that a biennial competition, Critical Assessment of Protein Structure Prediction (CASP), was launched to promote computational methods of solving the enigma. In 2020, Alphabet’s DeepMind entered the second iteration of their tool ‘AlphaFold’ into CASP14 and achieved such great success that Andriy Kryshtafovych, a project Scientist at UC Davis, declared the protein folding mystery “largely solved”.
AlphaFold 2 uses a machine learning approach to assess how a protein spontaneously organises its structure by analysing its amino acid chain. The algorithm is trained using a data pool of over 365,000 pre-sequenced proteins to create accurate predictions of stable protein structures when given their monomer sequence.
Govinda Bhisetti is a Principal Investigator and Head of Computational Chemistry at Biogen, who has a long-standing interest in protein structure and its application in drug design. Oxford Global hosted a discussion group led by Bhisetti in which the leading experts discussed what improvements they thought AlphaFold 2 would bring to the field of computational drug design.
The Success of AlphaFold 2
Bhisetti began the discussion by explaining his fascination with the success of AlphaFold. It is only recently that we are beginning to see the actual applications of machine learning in the real world. What was once confined to science fiction is now making its way into reality.
AlphaFold 2 has proved the most accurate protein structure prediction method by far. At CASP14, DeepMind’s project was over 2.5 times more accurate than the next best contestant (see figure. 1).
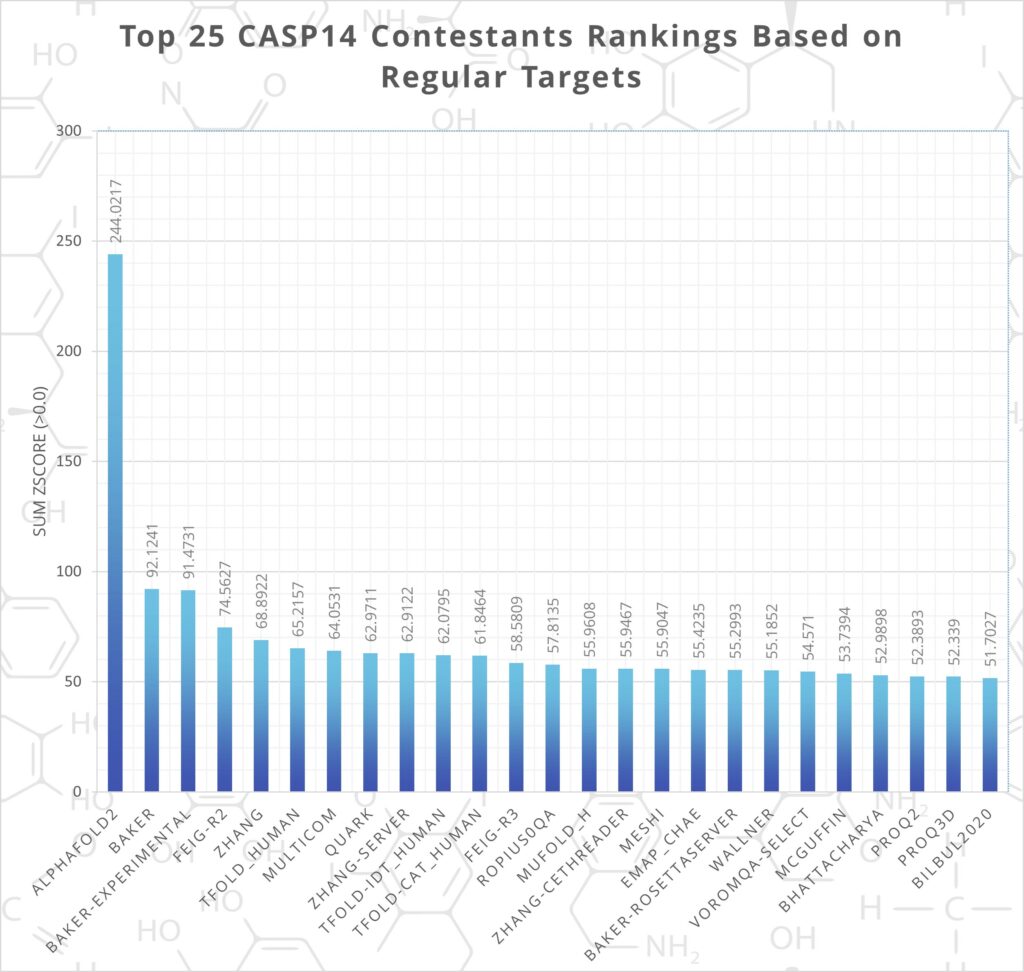
Figure 1. - CASP14 Rankings, AlphaFold2 Shown as Most Successful. Data from: predictioncenter.org/casp14/
Bhisetti then asked our attendees what they thought were the most important practical applications of the technology. The experts’ consensus hailed AlphaFold2 as a valuable technology with a bright future. However, its application is still rather in its infancy, mostly requiring combined approaches to make it worthwhile.
What Does AlphaFold Mean for Those in the Industry?
“[AlphaFold] is especially useful to optimise the binding mode of the ligand on the protein surface,” said Taiji Oashi, a Senior Scientist at Kyowa KIRIN. Oashi’s team is working on antibody structures and has had some success in using AlphaFold for predicting them. However, Oashi added that there are some differences in CDR H3 loop compared to data from X-ray crystallography, so there is still room for improvement for AlphaFold’s predictions.
It would be of great use and importance if AlphaFold were to improve toward predicting protein–ligand binding. But still, Oashi is sceptical about whether an AI approach would be the correct method for that kind of prediction. However, they have found a combined approach effective when working with proteins and their structures.
Sam Sinai is a Co-founder and Machine Learning Research Lead at Dyno Therapeutics, which is working on engineering a multimeric 60-monomer protein complex to bind to unknown receptors. Although Sinai’s research does not primarily rely on structure models his team uses AlphaFold to investigate regions of their protein in which there is no good crystal structure available.
They use the machine learning tool to see if their modifications to the protein maintain a stable structure. Sinai gave the example of a PLA domain in an AAV they are working on, which is not easily crystalised. His team are using AlphaFold predictions to understand whether the changes they make to its sequence will affect the structure of the protein.
Considering the future of the technology, Sinai commented that “using a machine learning tool like AlphaFold to predict protein–protein binding would be great, but we are far off from being able to reliably address that challenge for a complex multimeric protein complex and potential receptors.” He also noted the need to convince collaborators that it would be a good investment of time and resources.
Currently, Sinai’s company measures the protein function using high throughput technology after they edit the protein’s sequence: “It is still more cost-effective to measure the effects of modifications in-vitro” Sinai explained that AlphaFold, is fairly incremental in its impact on a multi-faceted problem such as gene delivery.
Limitations
Another hurdle is that AlphaFold is trained on already folded structures, which means that it could be too optimistic in folding sequences that are broken. “Sometimes you make a few changes to the sequence, and the protein does not fold properly anymore, but AlphaFold is not telling you that,” Sinai added. “Rather, it could approximate the protein back to whatever the closest structure is,” he continued. It is unclear if AlphaFold would give you too many false positives to be heavily relied upon for structural predictions at the moment.
The potential of AlphaFold—for the time being—is real, but it hasn’t been fully realised yet. Machine learning has the potential to revolutionise how scientists work with the structures of proteins. However, this can make it easy to overlook the fact that the technology is still in its early stages.
Our discussion group members think it will take some time before the computing power and cost-efficiency catches up to make AlphaFold the powerful tool it has the potential to be. However, they are still optimistic about the technology's future.
Got some insights to share? Come to our next discussion group which will be focussing on Oral PROTACs and PD/PK Profiling and will be taking place on Friday 29 April 2022 | 15:00 UTC+1. Register here >>
At Discovery Europe: In-Person, join and network with over 400 industry leaders at the renowned Drug Discovery Summit in Berlin, where we will address the latest advancements in target identification, validation and HIT optimisation.




.png)

